Initialising ...
Initialising ...
Initialising ...
Initialising ...
Initialising ...
Initialising ...
Initialising ...
Kawagoe, Soichiro*; Nakagawa, Hiroshi; Kumeta, Hiroyuki*; Ishimori, Koichiro*; Saio, Tomohide*
Journal of Biological Chemistry, 293(39), p.15095 - 15106, 2018/09
Times Cited Count:14 Percentile:49.8(Biochemistry & Molecular Biology)Molecular chaperones often possess functional modules that are specialized in assisting the formation of specific structural elements, such as a disulfide bridges and peptidyl-prolyl bonds in cis form, in the client protein. A ribosome-associated molecular chaperone Trigger Factor (TF), which has a peptidyl-prolyl cis/trans isomerase (PPIase) domain (PPD), acts as a highly efficient catalyst in the folding process limited by peptidyl-prolyl isomerization. Herein we report a study on the mechanism through which TF recognizes the proline residue in the unfolded client protein during the cis/trans isomerization process. The solution structure of TF in complex with the client protein showed that TF recognizes the proline-aromatic motif located in the hydrophobic stretch of the unfolded client protein through its conserved hydrophobic cleft, which suggests that TF preferentially accelerates the isomerization of the peptidyl-prolyl bond that is eventually folded into the core of the protein in its native fold. Molecular dynamics simulation revealed that TF exploits the backbone amide group of I195 to form an intermolecular hydrogen bond with the carbonyl oxygen of the amino acid residue preceding the proline residue at the transition state, which presumably stabilizes the transition state and thus accelerates the isomerization. The importance of such intermolecular hydrogen bond formation during the catalysis was further corroborated by the activity assay and NMR relaxation analysis.
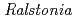 sp. A-471 reveals a unique arrangement of the catalytic residues for inverting chitin hydrolysis
sp. A-471 reveals a unique arrangement of the catalytic residues for inverting chitin hydrolysisArimori, Takao*; Kawamoto, Noriko*; Shinya, Shoko*; Okazaki, Nobuo*; Nakazawa, Masami*; Miyatake, Kazutaka*; Fukamizo, Tamo*; Ueda, Mitsuhiro*; Tamada, Taro
Journal of Biological Chemistry, 288(26), p.18696 - 18706, 2013/07
Times Cited Count:31 Percentile:64.14(Biochemistry & Molecular Biology)Chitinase C from 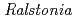 sp. A-471 (Ra-ChiC) has a catalytic domain sequence similar to goose type (G-type) lysozymes and, unlike other chitinases, belongs to glycohydrolase (GH) family 23. Using NMR spectroscopy, however, Ra-ChiC was found to interact only with the chitin dimer but not with the peptideglycan fragment. Here we report the crystal structures of wild-type, E141Q, and E162Q of the catalytic domain of Ra-ChiC with or without chitin oligosaccharides. Ra-ChiC has a substrate-binding site including a tunnel-shaped cavity, which determines the substrate specificity. Mutation analyses based on this structural information indicated that a highly conserved Glu141 acts as a catalytic acid, and that Asp226 located at the roof of the tunnel activates a water molecule as a catalytic base. The unique arrangement of the catalytic residues makes a clear contrast to the other GH23 members and also to inverting GH19 chitinases.
sp. A-471 (Ra-ChiC) has a catalytic domain sequence similar to goose type (G-type) lysozymes and, unlike other chitinases, belongs to glycohydrolase (GH) family 23. Using NMR spectroscopy, however, Ra-ChiC was found to interact only with the chitin dimer but not with the peptideglycan fragment. Here we report the crystal structures of wild-type, E141Q, and E162Q of the catalytic domain of Ra-ChiC with or without chitin oligosaccharides. Ra-ChiC has a substrate-binding site including a tunnel-shaped cavity, which determines the substrate specificity. Mutation analyses based on this structural information indicated that a highly conserved Glu141 acts as a catalytic acid, and that Asp226 located at the roof of the tunnel activates a water molecule as a catalytic base. The unique arrangement of the catalytic residues makes a clear contrast to the other GH23 members and also to inverting GH19 chitinases.
 -sheet-rich prion protein in scrapie-infected neuroblastoma cells with human IgG1 antibody specific for
-sheet-rich prion protein in scrapie-infected neuroblastoma cells with human IgG1 antibody specific for  -form prion protein
-form prion proteinKubota, Toshiya*; Hamazoe, Yuta*; Hashiguchi, Shuhei*; Ishibashi, Daisuke*; Akasaka, Kazuyuki*; Nishida, Noriyuki*; Katamine, Shigeru*; Sakaguchi, Suehiro*; Kuroki, Ryota; Nakashima, Toshihiro*; et al.
Journal of Biological Chemistry, 287(17), p.14023 - 14039, 2012/04
Times Cited Count:5 Percentile:11.59(Biochemistry & Molecular Biology)We prepared  -sheet-rich recombinant full-length prion protein (
-sheet-rich recombinant full-length prion protein ( -form PrP) (Jackson, G. S., Hosszu, L. L., Power, A., Hill, A. F., Kenney, J., Saibil, H., Craven, C. J., Waltho, J. P., Clarke, A. R., and Collinge, J. (1999) Science 283, 1935-1937). Using this
-form PrP) (Jackson, G. S., Hosszu, L. L., Power, A., Hill, A. F., Kenney, J., Saibil, H., Craven, C. J., Waltho, J. P., Clarke, A. R., and Collinge, J. (1999) Science 283, 1935-1937). Using this  -form PrP and a human single chain Fv-displaying phage library, we have established a human IgG1 antibody specific to
-form PrP and a human single chain Fv-displaying phage library, we have established a human IgG1 antibody specific to  -form but not
-form but not  -form PrP, PRB7 IgG. When prion-infected ScN2a cells were cultured with PRB7 IgG, they generated and accumulated PRB7-binding granules in the cytoplasm with time, consequently becoming apoptotic cells bearing very large PRB7-bound aggregates. The SAF32 antibody recognizing the N-terminal octarepeat region of full-length PrP stained distinct granules in these cells as determined by confocal laser microscopy observation. When the accumulation of proteinase K-resistant PrP was examined in prion-infected ScN2a cells cultured in the presence of PRB7 IgG or SAF32, it was strongly inhibited by SAF32 but not at all by PRB7 IgG. Thus, we demonstrated direct evidence of the generation and accumulation of
-form PrP, PRB7 IgG. When prion-infected ScN2a cells were cultured with PRB7 IgG, they generated and accumulated PRB7-binding granules in the cytoplasm with time, consequently becoming apoptotic cells bearing very large PRB7-bound aggregates. The SAF32 antibody recognizing the N-terminal octarepeat region of full-length PrP stained distinct granules in these cells as determined by confocal laser microscopy observation. When the accumulation of proteinase K-resistant PrP was examined in prion-infected ScN2a cells cultured in the presence of PRB7 IgG or SAF32, it was strongly inhibited by SAF32 but not at all by PRB7 IgG. Thus, we demonstrated direct evidence of the generation and accumulation of  -sheet-rich PrP in ScN2a cells de novo. These results suggest first that PRB7-bound PrP is not responsible for the accumulation of
-sheet-rich PrP in ScN2a cells de novo. These results suggest first that PRB7-bound PrP is not responsible for the accumulation of  -form PrP aggregates, which are rather an end product resulting in the triggering of apoptotic cell death, and second that SAF32-bound PrP lacking the PRB7-recognizing
-form PrP aggregates, which are rather an end product resulting in the triggering of apoptotic cell death, and second that SAF32-bound PrP lacking the PRB7-recognizing  -form may represent so-called PrPSc with prion propagation activity. PRB7 is the first human antibody specific to
-form may represent so-called PrPSc with prion propagation activity. PRB7 is the first human antibody specific to  -form PrP and has become a powerful tool for the characterization of the biochemical nature of prion and its pathology.
-form PrP and has become a powerful tool for the characterization of the biochemical nature of prion and its pathology.
Ito, Takachika*; Suzuki, Shoichi*; Kanaji, Sachiko*; Shiraishi, Hiroshi*; Ota, Shoichiro*; Arima, Kazuhiko*; Tanaka, Go*; Tamada, Taro; Honjo, Eijiro*; Garcia, K. C.*; et al.
Journal of Biological Chemistry, 284(36), p.24289 - 24296, 2009/09
Times Cited Count:24 Percentile:45.24(Biochemistry & Molecular Biology)Both IL-4 and IL-13 can bind to the shared receptor composed of the IL-4 receptor  chain and the IL-13 receptor
chain and the IL-13 receptor  -1 chain (IL-13R
-1 chain (IL-13R 1); however, the assembly mechanisms of these ligands to the receptor is different, enabling the principal functions of these ligands to be different. We have previously shown that the N-terminal Ig-like domain in IL-13R
1); however, the assembly mechanisms of these ligands to the receptor is different, enabling the principal functions of these ligands to be different. We have previously shown that the N-terminal Ig-like domain in IL-13R 1, called the D1 domain, is the specific and critical binding unit for IL-13. However, it has still remained obscure which the amino acid has specific binding capacity to IL-13 and why the D1 domain acts as the binding site for IL-13, but not IL-4. To address these questions, in this study, we performed the mutational analyses for the D1 domain, combining the structural data to identify the amino acids critical for binding to IL-13. Mutations of Lys76, Lys77, or Ile78 in c' strand in which the crystal structure showed interact with IL-13 and those of Trp65 and Ala79 adjacent to the interacting site, resulted in significant impairment of IL-13 binding, demonstrating that these amino acids generate the binding site. Furthermore, mutations of Val35, Leu38, or Val42 at N-terminal
1, called the D1 domain, is the specific and critical binding unit for IL-13. However, it has still remained obscure which the amino acid has specific binding capacity to IL-13 and why the D1 domain acts as the binding site for IL-13, but not IL-4. To address these questions, in this study, we performed the mutational analyses for the D1 domain, combining the structural data to identify the amino acids critical for binding to IL-13. Mutations of Lys76, Lys77, or Ile78 in c' strand in which the crystal structure showed interact with IL-13 and those of Trp65 and Ala79 adjacent to the interacting site, resulted in significant impairment of IL-13 binding, demonstrating that these amino acids generate the binding site. Furthermore, mutations of Val35, Leu38, or Val42 at N-terminal  -strand also resulted in loss of IL-13 binding, probably from decrease structural stability. None of the mutations employed here affected IL-4 binding. These results demonstrate that the hydrophobic patch composed of Lys76, Lys77, and Ile78 is the IL-13 recognition site and solidify our understanding that the differential requirements of the D1 domain in IL-13R
-strand also resulted in loss of IL-13 binding, probably from decrease structural stability. None of the mutations employed here affected IL-4 binding. These results demonstrate that the hydrophobic patch composed of Lys76, Lys77, and Ile78 is the IL-13 recognition site and solidify our understanding that the differential requirements of the D1 domain in IL-13R 1 allows the shared receptor to respond differentially to IL-4 and IL-13.
1 allows the shared receptor to respond differentially to IL-4 and IL-13.